Antimicrobial Activities of Sponge-Derived Microorganisms from Coastal Waters of Central Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Sponge-Associated Bacteria-SAB and Test Strain Vibrio sp.
2.3. Screening of Strains for PKS and NRPSs
2.4. Primary Screening of Antimicrobial Activities by Colony Picking Method
2.5. Antimicrobial Activity Testing by Well Diffusion Method
2.6. Identification of Antimicrobial Producing Strains
2.7. Data Analysis
3. Results
3.1. Sponge Associated Bacteria with PKS- and NRPS-Holders
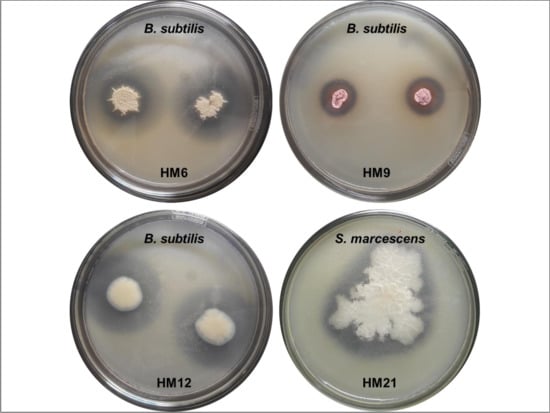
3.2. Antimicrobial Tests
3.3. Identification of Potential Strains.
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Love, G.D.; Grosjean, E.; Stalvies, C.; Fike, D.A.; Grotzinger, J.P.; Bradley, A.S.; Kelly, A.E.; Bhatia, M.; Meredith, W.; Snape, C.E.; et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 2009, 457, 718–721. [Google Scholar] [CrossRef] [Green Version]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef] [Green Version]
- Aleti, G.; Sessitsch, A.; Brader, G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput. Struct. Biotechnol. J. 2015, 13, 192–203. [Google Scholar] [CrossRef]
- Zhao, X.; Kuipers, O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016, 17, 882. [Google Scholar] [CrossRef] [Green Version]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Codd, R. Copper(II)-based metal affinity chromatography for the isolation of the anticancer agent bleomycin from Streptomyces verticillus culture. J. Inorg. Biochem. 2012, 115, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.T.; Komaki, H.; Motohashi, K.; Kozone, I.; Mukai, A.; Takagi, M.; Shin-ya, K. Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products. Environ. Microbiol. 2011, 13, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Kanagasabhapathy, M.; Janussen, D.; Xue, S.; Zhang, W. Phylogenetic diversity of Gram-positive bacteria cultured from Antarctic deep-sea sponges. Polar Biol. 2011, 34, 1501–1512. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef]
- Leal, M.C.; Sheridan, C.; Osinga, R.; Dionisio, G.; Rocha, R.J.; Silva, B.; Rosa, R.; Calado, R. Marine microorganism-invertebrate assemblages: Perspectives to solve the “supply problem” in the initial steps of drug discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indraningrat, A.A.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, J.A. Diversity and biotechnological potential of microorganisms associated with marine sponges. Appl. Microbiol. Biotechnol. 2014, 98, 7331–7347. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Natural Products from Marine Fungi—Still an Underrepresented Resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef]
- Do, T.T.; Duong, T.D.; Nguyen, X.N.; Bui, H.T.; Pham, H.Y.; Hoang, L.T.A.; Do, C.T.; Chau, V.M.; Phan, V.K. Furanosesterterpenes from the marine sponge Ircinia echinata (Keller, 1889). Vietnam J. Chem. 2016, 54, 477. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Chau, V.M.; Tran, T.H.; Phan, V.K.; Hoang, T.H.; Nguyen, T.D.; Nguyen, X.N.; Tai, B.H.; Hyun, J.H.; Kang, H.K.; et al. C29 sterols with a cyclopropane ring at C-25 and 26 from the Vietnamese marine sponge Ianthella sp. and their anticancer properties. Bioorg. Med. Chem. Lett. 2009, 19, 4584–4588. [Google Scholar] [CrossRef]
- Trinh, T.T.V.; Truong, B.N.; Longeon, A.; Doan, T.M.H.; Deville, A.; Chau, V.M.; Pham, V.C.; Bourguet-Kondracki, M.L. New 9alpha-Hydroxy-5alpha,6alpha-epoxyhydroxysterols from the Vietnamese Marine Sponge Ircinia echinata. Mar. Drugs 2018, 16, 424. [Google Scholar] [CrossRef] [Green Version]
- Ton, T.H.D.; Steinert, G.; Nguyen, T.K.C.; Smidt, H.; Sipkema, D. Archaeal and bacterial diversity and community composition from 18 phylogenetically divergent sponge species in Vietnam. PeerJ 2018, 6, e4970. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.-C.; Putchakarn, S.; Thai, M.-Q.; Wang, D.; Huang, Y.M. Inventory of sponge fauna from the Singapore Strait to Taiwan Strait along the western coastline of the South China Sea. Raffles Bull. Zool. 2016, 34, 104–129. [Google Scholar]
- Hettiarachchi, S.; Lee, S.J.; Lee, Y.; Kwon, Y.K.; De Zoysa, M.; Moon, S.; Jo, E.; Kim, T.; Kang, D.H.; Heo, S.J.; et al. A Rapid and Efficient Screening Method for Antibacterial Compound-Producing Bacteria. J. Microbiol. Biotechnol. 2017, 27, 1441–1448. [Google Scholar] [CrossRef]
- Ayuso-Sacido, A.; Genilloud, O. New PCR Primers for the Screening of NRPS and PKS-I Systems in Actinomycetes: Detection and Distribution of These Biosynthetic Gene Sequences in Major Taxonomic Groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Metsä-Ketelä, M.; Salo, V.; Halo, L.; Hautala, A.; Hakala, J.; Mäntsälä, P.; Ylihonko, K. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 1999, 180, 1–6. [Google Scholar] [CrossRef]
- Woese, C.R.; Gutell, R.; Gupta, R.; Noller, H.F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol. Rev. 1983, 47, 621–669. [Google Scholar] [CrossRef] [PubMed]
- Wikler, M.A.; Hindler, J.F.; Cockerill, F.R.; Patel, J.B.; Bush, K.; Powell, M.; Dudley, M.N.; Turnidge, J.D.; Eliopoulos, G.M.; Weinstein, M.P.; et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard—Eighth Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009; Volume 29. [Google Scholar]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathog. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Pham, T.M.; Wiese, J.; Wenzel-Storjohann, A.; Imhoff, J.F. Diversity and antimicrobial potential of bacterial isolates associated with the soft coral Alcyonium digitatum from the Baltic Sea. Anton. Leeuw. 2016, 109, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Tian, Y.; Niu, G.; Tan, H. GouR, a TetR family transcriptional regulator, coordinates the biosynthesis and export of gougerotin in Streptomyces graminearus. Appl. Environ. Microbiol. 2014, 80, 714–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, T.M. Community of Soft Coral Alcyonium Digitatum Associated Bacteria and Their Antimicrobial Activities. Ph.D. Thesis, Christian-Albrecht University of Kiel, GEOMAR, Kiel, Germany, 2014. [Google Scholar]
- Wiese, J.; Abdelmohsen, U.R.; Motiei, A.; Humeida, U.H.; Imhoff, J.F. Bacicyclin, a new antibacterial cyclic hexapeptide from Bacillus sp. strain BC028 isolated from Mytilus edulis. Bioorg. Med. Chem. Lett. 2018, 28, 558–561. [Google Scholar] [CrossRef]
- Pham, V.C.; Nguyen, M.A.; Vu, T.Q.; Nguyen, T.K.C. Isolation, screening and identification of some sponge associated bacterial isolates from six marine sponge species of Son Cha coast. Vietnam J. Biol. 2015, 36, 345–350. [Google Scholar]
- Pham, T.M.; Nguyen, N.T.; Nguyen, K.H. Bacillus sp. VK2 isolated from Acropora hyacinthus from Ninh Thuan and its antimicrobial activities against cause of white pox disease in Acropora palmate. Vietnam J. Mar. Sci. Technol. 2018, 18, 197–204. [Google Scholar]
- Pham, T.M.; Dao, V.H.; Nguyen, K.H. Antibiotic resistance of marine bacteria from Mot Island Nha Trang Bay. Vietnam J. Mar. Sci. Technol. 2017, 17, 466–475. [Google Scholar]
- Thiel, V.; Leininger, S.; Schmaljohann, R.; Brümmer, F.; Imhoff, J.F. Sponge-specific Bacterial Associations of the Mediterranean Sponge Chondrilla nucula (Demospongiae, Tetractinomorpha). Microb. Ecol. 2007, 54, 101–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, S.; Suvathi, S.S.; Aysha, O.S.; Nirmala, P.; Vinoth, K.P.; Reena, A. Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian Pac. J. Trop. Biomed. 2012, 2, 469–473. [Google Scholar] [CrossRef] [Green Version]
- Arifuzzaman, M.; Khatun, M.R.; Rahman, H. Isolation and screening of actinomycetes from Sundarbans soil for antibacterial activity. Afr. J. Biotechnol. 2010, 9, 4615–4619. [Google Scholar]
- Ceylan, O.; Okmen, G.; Ugur, A. Isolation of soil Streptomyces as source antibiotics active against antibiotic-resistant bacteria. Eurasian J. Biosci. 2008, 2, 73–82. [Google Scholar]
- Pham, T.M.; Nguyen, K.H.; Nguyen, M.H.; Phan, M.T.; Hoang, T.D.; Vo, H.T.; Nguyen, T.D.H.; Le, T.D.; Nguyen, H.H. A study on bacteria associated with three hard coral species from Ninh Thuan by epifluorescence and most diluted culture method. J. Mar. Sci. Technol. 2019, 19, 271–283. [Google Scholar]
- Nguyen, K.H.; Pham, T.M.; Bui, H.H.; Vo, H.T. Screening of coral associated bacteria with antimicrobial activities from scleractinian coral Acropora muricata in the Nha Trang bay. Collect. Mar. Work 2016, 22, 83–95. [Google Scholar]
- Patterson, K.L.; Porter, J.W.; Ritchie, K.B.; Polson, S.W.; Mueller, E.; Peters, E.C.; Santavy, D.L.; Smith, G.W. The Etiology of White Pox, A Lethal Disease of the Caribbean Elkhorn Coral, Acropora palmata. Proc. Natl. Acad. Sci. USA 2002, 99, 8725–8730. [Google Scholar] [CrossRef] [Green Version]






| Isolates | Colony Color | Colony Shapes and Sizes after 48 h on R2A Medium | Genes Holder |
|---|---|---|---|
| HM1 | Off-white | Round, concentric circle, convex surface. CZ 3–4. | ND |
| HM2 | Opaque | Round, smooth wet surface. CZ 3–4. | NRPS |
| HM3 | Off-white | Round, convex surface. CZ 3–4. | ND |
| HM4 | Opaque | Circular. Punctiform colonies. | ND |
| HM5 | Off-white | Round, convex concentric. Rod cell shape. CZ 1–2. | NRPS PKSII |
| HM6 | Off-white | Circular, raised smooth surface. Rod cell shape. CZ 3–5. | NRPS PKSII |
| HM7 | Milky-white | Dry surface lobes, irregular form. CZ 3–5. | ND |
| HM8 | Milky-white | Circular, raised, smooth, curled. Rod cell shape. CZ 5–7. | ND |
| HM9 | Reddish | Round, convex, concentric circle, white layer like chalk dust. Network long rod cell. CZ 1–2. | NRPS PKSII |
| HM10 | White | Round, smooth surface. Punctiform. | ND |
| HM11 | Brown | Lobe form, smooth wet surface. CZ 2–3. | ND |
| HM12 | Off-white | Circular, smooth, raised. Coccus cell. Punctiform. | PKSII |
| HM13 | Milky-white | Round, convex, wet lobe surface. CZ 1–2. | ND |
| HM14 | Bright | Round, convex, iridescent surfaces. CZ 3–4. | ND |
| HM15 | Off-white | Round, jagged edges, pale protruding center. CZ 3–4. | ND |
| HM16 | Off-white | Round colonies, rough surface. CZ 2–3 | ND |
| HM17 | Milky-white | Round, dry edges, lobed border. CZ 4–5 | ND |
| HM18 | Milky-white | Round, smooth. CZ 1–2. | ND |
| HM19 | Brown | Round, convex iridescent, wet surface.CZ 1–2. | ND |
| HM20 | Yellow | Round, convex surface. CZ 2–3. | ND |
| HM21 | Milky-white | Round, convex, darkest center. CZ 4–5. | NRPS PKSII |
| HM22 | Milky-white | Radioactive colonies, scald surface, lobes. Punctiform. | NRPS |
| HM23 | Yellow | Round, flat, glossy surface. CZ 2–4. | ND |
| HM24 | Yellow | Round, smooth wet convex surface. Punctiform. | ND |
| HM25 | Opaque | Round form, smooth wet convex surface. CZ 2–4. | ND |
| Test Strains | Holder PKS/NRPS Strains | ||||||
|---|---|---|---|---|---|---|---|
| HM2 | HM5 | HM6 | HM9 | HM12 | HM21 | HM22 | |
| B. subtilis | − | − | + | + | + | − | − |
| S. aureus | − | − | − | − | − | − | − |
| E. coli | − | − | − | − | − | − | − |
| S. typhimurium | − | − | − | − | − | − | − |
| S. marcescens | − | − | − | − | − | + | − |
| C. albicans | − | − | − | − | − | − | − |
| V. parahaemolyticus | − | − | − | − | − | − | − |
| V. campbellii | − | − | − | − | − | − | − |
| Isolate Code | NRPS/PKS | Colony Picking | Well Diffusion | Next Related Type Strains (RDPII) | Possible Genus |
|---|---|---|---|---|---|
| HM2 | NRPS | ND | A | Bacillus amyloliquefaciensT FZB42 (CP000560: 99.6%) | Bacillus sp. |
| HM5 | NRPS PKSII | ND | A, B | Bacillus amyloliquefaciensT FZB42 (CP000560: 98.1%) | Bacillus sp. |
| HM6 | NRPS PKSII | A | A, B, C, D | Bacillus amyloliquefaciensT FZB42 (CP000560: 99.6%) | Bacillus sp. |
| HM8 | ND | ND | A, B | Bacillus subtilisT DSM22148 (HE582781: 99.0%) | Bacillus sp. |
| HM9 | NRPS PKSII | A | A, B, C, E, F | Streptomyces mexicanusT NBRC 100915 (AB249966: 94%) | Streptomyces sp. |
| HM12 | PKSII | A | A, C | Bacillus subtilisT DSM 22148 (HE582781: 99.5%) | Bacillus sp. |
| HM19 | ND | ND | B, C, D | Bacillus toyonensisT CNCM I-1012 (AJ310100: 99.8%) | Bacillus sp. |
| HM20 | ND | ND | A, B, C, D | Bacillus safensisT FO-036b (AF234854: 99.6%) | Bacillus sp. |
| HM21 | NRPS PKSII | C | A | Bacillus amyloliquefaciensT FZB42 (CP000560: 99.6%) | Bacillus sp. |
| HM22 | NRPS | ND | C | Bacillus safensisT FO-036b (AF234854: 100%) | Bacillus sp. |
| HM23 | ND | ND | A | Bacillus cereusT ATCC 14579 (AE016877: 99.6%) | Bacillus sp. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mien, P.T.; Ha, D.V.; Ben, H.X.; Chen, B.; Liu, L.; Minh-Thu, P. Antimicrobial Activities of Sponge-Derived Microorganisms from Coastal Waters of Central Vietnam. J. Mar. Sci. Eng. 2020, 8, 594. https://0-doi-org.brum.beds.ac.uk/10.3390/jmse8080594
Mien PT, Ha DV, Ben HX, Chen B, Liu L, Minh-Thu P. Antimicrobial Activities of Sponge-Derived Microorganisms from Coastal Waters of Central Vietnam. Journal of Marine Science and Engineering. 2020; 8(8):594. https://0-doi-org.brum.beds.ac.uk/10.3390/jmse8080594
Chicago/Turabian StyleMien, Pham Thi, Dao Viet Ha, Hoang Xuan Ben, Bin Chen, Lan Liu, and Phan Minh-Thu. 2020. "Antimicrobial Activities of Sponge-Derived Microorganisms from Coastal Waters of Central Vietnam" Journal of Marine Science and Engineering 8, no. 8: 594. https://0-doi-org.brum.beds.ac.uk/10.3390/jmse8080594