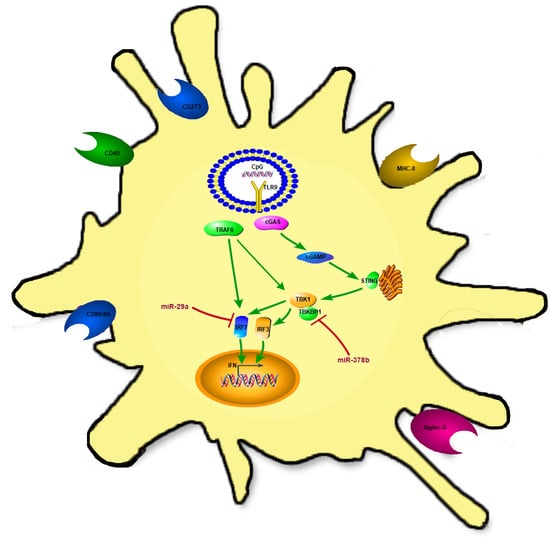
miR29a and miR378b Influence CpG-Stimulated Dendritic Cells and Regulate cGAS/STING Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Ethical Statement
2.2. miRNA Selection and Quantification
2.3. In Silico miRNA Target Site Prediction
2.4. Cell Isolation, Culture, and Surface Marker Detection
2.5. Plasmids and miRNA Inhibitors
2.6. Reagents and Antibodies
2.7. Quantitative Real-Time Reverse Transcriptase PCR
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. Western Blotting Analysis
2.10. Dual Luciferase Assay
2.11. Statistical Analysis
3. Results
3.1. CpG Influences miRNAs Level in Bone Marrow-Derived Dendritic Cells (BMDCs)
3.2. miR-29a and miR-378b Regulate Co-Stimulatory Molecules of BMDCs
3.3. miR-29a and miR-378b Modulate the Immunosuppressive Molecules CD273 and Siglec-G in DCs
3.4. miR-29a and miR-378b Influence the cGAS/STING Pathway
3.4.1. mRNA Analysis of cGAS/STING Pathway Changes Stimulated by miR-29a and miR-378b
3.4.2. cGAS/STING Pathway Regulatory Proteins Influenced by miR-29a and miR-378b
3.5. miR-29a and miR-378b Infliuence Mitogen-Activated Protein Kinase (MAPK) and TRAF6 Pathways in CpG-Stimulated BMDCs
3.6. CpG-Induced miRNAs Influence Cytokine Secretion by DCs
3.7. miR-29a and miR-378b Target Prediction and Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DCs | dendritic cells |
| miRNA | microRNA |
| CpG | Cytosine–phosphate–guanosine |
| BMDC | bone marrow dendritic cells |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| MHCII | major histocompatibility complex class II |
| Siglec-G | sialic acid-binding immunoglobulin-type lectin-G |
| 3′UTR | three-prime untranslated region |
| FACS | Fluorescence-Activated Cell Sorter |
| MAPK | mitogen-activated protein kinases |
| cGAS/STING | cyclic GMP–AMP/synthase-stimulator of interferon genes |
| TBKBP1 | TANK-binding kinase binding protein 1 |
| IRF7 | interferon regulatory factor 7 |
| TLR-9 | Toll-like receptor 9 |
References
- Arnold-Schrauf, C.; Dudek, M.; Dielmann, A.; Pace, L.; Swallow, M.; Kruse, F.; Kuhl, A.A.; Holzmann, B.; Berod, L.; Sparwasser, T. Dendritic cells coordinate innate immunity via MyD88 signaling to control Listeria monocytogenes infection. Cell Rep. 2014, 6, 698–708. [Google Scholar] [CrossRef]
- Ding, Y.; Guo, Z.; Liu, Y.; Li, X.; Zhang, Q.; Xu, X.; Gu, Y.; Zhang, Y.; Zhao, D.; Cao, X. The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat. Immunol. 2016, 17, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Scheiermann, J.; Klinman, D.M. Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 2014, 32, 6377–6389. [Google Scholar] [CrossRef] [PubMed]
- Bode, C.; Zhao, G.; Steinhagen, F.; Kinjo, T.; Klinman, D.M. CpG DNA as a vaccine adjuvant. Expert Rev. Vaccines 2011, 10, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002, 20, 709–760. [Google Scholar] [CrossRef]
- Wagner, H. Interactions between bacterial CpG-DNA and TLR9 bridge innate and adaptive immunity. Curr. Opin. Microbiol. 2002, 5, 62–69. [Google Scholar] [CrossRef]
- Putta, M.; Zhu, F.-G.; Wang, D.; Bhagat, L.; Dai, M.; Kandimalla, E.; Agrawal, S. Peptide Conjugation at the 5′-End of Oligodeoxynucleotides Abrogates Toll-Like Receptor 9-Mediated Immune Stimulatory Activity. Bioconjug. Chem. 2009, 21, 39–45. [Google Scholar] [CrossRef]
- Vollmer, J.; Krieg, A.M. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv. Drug Deliv. Rev. 2009, 61, 195–204. [Google Scholar] [CrossRef]
- Kadowaki, N.; Antonenko, S.; Liu, Y.J. Distinct CpG DNA and Polyinosinic-Polycytidylic Acid Double-Stranded RNA, Respectively, Stimulate CD11c- Type 2 Dendritic Cell Precursors and CD11c+ Dendritic Cells to Produce Type I IFN. J. Immunol. 2001, 166, 2291–2295. [Google Scholar] [CrossRef]
- Klinman, D.M. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 2004, 4, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, C.; Yue, R.; El-Ashram, S.; Wang, J.; He, X.; Zhao, D.; Zhou, X.; Xu, L. cGAS/STING/TBK1/IRF3 Signaling Pathway Activates BMDCs Maturation Following Mycobacterium bovis Infection. Int. J. Mol. Sci. 2019, 20, 895. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Y.Y.; Lu, J.S.; Xia, B.H.; Yang, Z.X.; Zhu, X.D.; Zhou, X.W.; Huang, P.T. The highly pathogenic H5N1 influenza A virus down-regulated several cellular MicroRNAs which target viral genome. J. Cell. Mol. Med. 2017, 21, 3076–3086. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, X.; Yu, B.; Chen, D. Cloning and functional characterization of rat stimulator of interferon genes (STING) regulated by miR-24. Dev. Comp. Immunol. 2012, 37, 414–420. [Google Scholar] [CrossRef]
- Ingle, H.; Kumar, S.; Raut, A.A.; Mishra, A.; Kulkarni, D.D.; Kameyama, T.; Takaoka, A.; Akira, S.; Kumar, H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015, 8. [Google Scholar] [CrossRef]
- Smyth, L.A.; Boardman, D.A.; Tung, S.L.; Lechler, R.; Lombardi, G. MicroRNAs affect dendritic cell function and phenotype. Immunology 2015, 144, 197–205. [Google Scholar] [CrossRef]
- Lin, J.; Yin, Y.Y.; Qin, T.; Zhu, L.Q.; Yu, Q.H.; Yang, Q. Enhanced immune response of BMDCs pulsed with H9N2 AIV and CpG. Vaccine 2014, 32, 6783–6790. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lin, J.; Xia, J.; Zhang, T.; Zhang, K.; Yang, Q. Genome-wide profiling of microRNAs reveals novel insights into the interactions between H9N2 avian influenza virus and avian dendritic cells. Oncogene 2018, 37, 4562–4580. [Google Scholar] [CrossRef]
- Halperin, S.A.; Van Nest, G.; Smith, B.; Abtahi, S.; Whiley, H.; Eiden, J.J. A phase I study of the safety and immunogenicity of recombinant hepatitis B surface antigen co-administered with an immunostimulatory phosphorothioate oligonucleotide adjuvant. Vaccine 2003, 21, 2461–2467. [Google Scholar] [CrossRef]
- Johanson, T.M.; Cmero, M.; Wettenhall, J.; Lew, A.M.; Zhan, Y.; Chong, M.M. A microRNA expression atlas of mouse dendritic cell development. Immunol. Cell Biol. 2015, 93, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Xia, J.; Tu, C.Z.; Zhang, K.Y.; Zeng, Y.; Yang, Q. H9N2 Avian Influenza Virus Protein PB1 Enhances the Immune Responses of Bone Marrow-Derived Dendritic Cells by Down-Regulating miR375. Front. Microbiol. 2017, 8, 287. [Google Scholar] [CrossRef]
- Abomaray, F.M.; Al Jumah, M.A.; Kalionis, B.; AlAskar, A.S.; Al Harthy, S.; Jawdat, D.; Al Khaldi, A.; Alkushi, A.; Knawy, B.A.; Abumaree, M.H. Human Chorionic Villous Mesenchymal Stem Cells Modify the Functions of Human Dendritic Cells, and Induce an Anti-Inflammatory Phenotype in CD1+ Dendritic Cells. Stem Cell Rev. Rep. 2015, 11, 423–441. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Huang, G.; Shi, L.Z.; Chi, H. Regulation of JNK and p38 MAPK in the immune system: Signal integration, propagation and termination. Cytokine 2009, 48, 161–169. [Google Scholar] [CrossRef]
- Liu, Y.; Shepherd, E.G.; Nelin, L.D. MAPK phosphatases—Regulating the immune response. Nat. Rev. Immunol. 2007, 7, 202–212. [Google Scholar] [CrossRef]
- Zhao, T.; Yang, L.; Sun, Q.; Arguello, M.; Ballard, D.W.; Hiscott, J.; Lin, R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat. Immunol. 2007, 8, 592–600. [Google Scholar] [CrossRef]
- Chariot, A.; Leonardi, A.; Muller, J.; Bonif, M.; Brown, K.; Siebenlist, U. Association of the adaptor TANK with the I kappa B kinase (IKK) regulator NEMO connects IKK complexes with IKK epsilon and TBK1 kinases. J. Biol. Chem. 2002, 277, 37029–37036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinho, F.V.; Benmerzoug, S.; Rose, S.; Campos, P.C.; Marques, J.T.; Bafica, A.; Barber, G.; Ryffel, B.; Oliveira, S.C.; Quesniaux, V.F.J. The cGAS/STING Pathway Is Important for Dendritic Cell Activation but Is Not Essential to Induce Protective Immunity against Mycobacterium tuberculosis Infection. J. Innate Immun. 2018, 10, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, Y.; Juma, C.A.; Yang, C.; Huang, J.; Zhang, X.; Zeng, Y. Differential Inhibition of Target Gene Expression by Human microRNAs. Cells 2019, 8, 791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschberger, S.; Hinske, L.C.; Kreth, S. MiRNAs: Dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018, 431, 11–21. [Google Scholar] [CrossRef]
- Toubai, T.; Rossi, C.; Oravecz-Wilson, K.; Zajac, C.; Liu, C.; Braun, T.; Fujiwara, H.; Wu, J.; Sun, Y.; Brabbs, S.; et al. Siglec-G represses DAMP-mediated effects on T cells. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Han, C.; Xie, B.; Hu, X.; Yu, Q.; Shi, L.; Wang, Q.; Li, D.; Wang, J.; Zheng, P.; et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 2013, 152, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Xu, S.; Liu, X.; Zhang, Q.; Xu, X.; Liu, M.; Hua, M.; Li, N.; Yao, H.; Cao, X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 2011, 12, 861–869. [Google Scholar] [CrossRef]
- Joo, D.; An, S.; Choi, B.G.; Kim, K.; Choi, Y.M.; Ahn, K.J.; An, I.S.; Cha, H.J. MicroRNA378b regulates alpha1type 1 collagen expression via sirtuin 6 interference. Mol. Med. Rep. 2017, 16, 8520–8524. [Google Scholar] [CrossRef] [Green Version]
- Deng, H.; Guo, Y.; Song, H.; Xiao, B.; Sun, W.; Liu, Z.; Yu, X.; Xia, T.; Cui, L.; Guo, J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 2013, 518, 351–359. [Google Scholar] [CrossRef]
- Hyun, J.; Wang, S.; Kim, J.; Rao, K.M.; Park, S.Y.; Chung, I.; Ha, C.S.; Kim, S.W.; Yun, Y.H.; Jung, Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat. Commun. 2016, 7, 10993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.Y.; Deng, Z.; Wang, C.H.; Yang, B.B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl. Acad. Sci. USA 2007, 104, 20350–20355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, L.; Gu, W.; Chen, L.; Pan, L.; Chen, J.; Peng, Y. MiR-378 overexpression attenuates high glucose-suppressed osteogenic differentiation through targeting CASP3 and activating PI3K-Akt signaling pathway. Int. J. Clin. Exp. Pathol. 2014, 7, 7249–7261. [Google Scholar] [PubMed]
- Wang, X.L.; Zhang, T.; Wang, J.; Zhang, D.B.; Zhao, F.; Lin, X.W.; Wang, Z.; Shi, P.; Pang, X.N. MiR-378b Promotes Differentiation of Keratinocytes through NKX3.1. PLoS ONE 2015, 10, e0136049. [Google Scholar] [CrossRef]
- Krist, B.; Florczyk, U.; Pietraszek-Gremplewicz, K.; Jozkowicz, A.; Dulak, J. The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int. J. Endocrinol. 2015, 2015, 281756. [Google Scholar] [CrossRef] [Green Version]
- Hua, C.; Sun, L.; Yang, Y.; Tan, R.; Hou, Y. Mechanisms of CpG-induced CD40 expression on murine bone marrow-derived dendritic cells. Autoimmunity 2013, 46, 177–187. [Google Scholar] [CrossRef]
- Bode, C.; Fox, M.; Tewary, P.; Steinhagen, A.; Ellerkmann, R.K.; Klinman, D.; Baumgarten, G.; Hornung, V.; Steinhagen, F. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur. J. Immunol. 2016, 46, 1615–1621. [Google Scholar] [CrossRef]
- Cai, X.; Chiu, Y.H.; Chen, Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Li, T.; Chen, Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Wu, M.Z.; Cheng, W.C.; Chen, S.F.; Nieh, S.; O’Connor, C.; Liu, C.L.; Tsai, W.W.; Wu, C.J.; Martin, L.; Lin, Y.S.; et al. miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat. Cell Biol. 2017, 19, 1286–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberger, C.M.; Podyminogin, R.L.; Diercks, A.H.; Treuting, P.M.; Peschon, J.J.; Rodriguez, D.; Gundapuneni, M.; Weiss, M.J.; Aderem, A. miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis. PLOS Pathogen 2017, 13, e1006305. [Google Scholar] [CrossRef] [PubMed]
- Nagalingam, R.S.; Sundaresan, N.R.; Gupta, M.P.; Geenen, D.L.; Solaro, R.J.; Gupta, M. A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J. Biol. Chem. 2013, 288, 11216–11232. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| miRNAs | Sense Primers (5′ to 3′) |
|---|---|
| mmu-miR-29a | AGAGGATGACTGATTTCTTTTGGTGTTC |
| mmu-miR-98 | GCTGGGGTGAGGTAGTAAGTTGTATTGT |
| mmu-miR-196a-2 | GTGGCTTAGGTAGTTTCATGTTGTTGG |
| mmu-miR-222 | AGTGGCTCAGTAGCCAGTGTAGATCC |
| mmu-miR-361 | GAAGCTTATCAGAATCTCCAGGGGTAC |
| mmu-miR-378b | GGGAACCCTGGACTTGGAGTCAGAAGA |
| miRNAs | Sequence | Products |
|---|---|---|
| miR-29a sense | GGCTCGAGACACCCACCATCACTATGTG | 461 bp |
| miR-29a anti-sense | TGAATTCAACAGGCTACCAGAGCCTG | |
| miR-378b sense | GGCTCGAGTATCATGGCATATCAGCAGAAGC | 488 bp |
| miR-378b anti-sense | TGAATTCTGCATATACCTGATAAGTCACAGG |
| Gene | Sequence | Products |
|---|---|---|
| mmu-miR-29a-5p target gene-Interferon Regulatory Factor 7 (IRF7) wild-type sense | AAGATCCTTTATTAAGCTTAAATCAGTGGAGCCCTGGGT | 406 bp |
| mmu-miR-29a-5p target gene-IRF7 wild-type anti-sense | GCACTAGTGAGGGAGCTCGGTTTCGGAAAGCCTGACGG | |
| mmu-miR-29a-5p target gene-IRF7 mutant-type sense | AAGATCCTTTATTAAGCTTGCTATCATGGAGCCCTGGGT | |
| mmu-miR-378b target gene-TANK-Binding Kinase Binding Protein 1 (TBKBP1) wild-type sense | GATCCTTTAAGCTTAAGTCCAATAAAACTTACCTG | 483 bp |
| mmu-miR-378b target gene-TBKBP1 wild-type anti-sense | CACTAGTGAGGGAGCTCAAAAGTCATGAGTTTGTGAC | |
| mmu-miR-378b target gene-TBKBP1 mutant type sense | GATCCTTTATTAAGCTTGGTGTAGGATAAAACTTACCTG |
| Gene | Sense | Anti-Sense |
|---|---|---|
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
| cGAS | CATGTGTGCAGGAGCATGTA | CAACAACCCATGCAACAAAG |
| STING | TGAGCCTCAACCAACCCTAC | CCATCCACACAGGTCAACAG |
| TBK1 | GGGGTGCTCTCCCTAATTCT | CTTGTCAGGGAACCGACTGT |
| TBKBP1 | GGTGAGGGGTATGTCAGCAG | TCCAAGACCTTCCGAGTGAG |
| IRF3 | CAAGGCTCAGTCTTCCCATC | CGTAGGGACAATGTGTGTGC |
| IRF7 | TCTCGGCTTGTGCTTGTCTA | ACTGGGGGTCACCTTCTTTC |
| STAT6 | TCTGCTTTTGCCAGTGTGAC | GCCCAGGGAGTTTACACAGA |
| TRAF6 | TGGGTCCTCTGTGTCTTTGA | AGGAGTCAGATGGGGCTACA |
| NEMO | GCTCCTGTTGTCTCCTTTGC | TGCAGTTTCCCTGTGTTGAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, A.U.; Cao, Y.; Siddique, N.; Lin, J.; Yang, Q. miR29a and miR378b Influence CpG-Stimulated Dendritic Cells and Regulate cGAS/STING Pathway. Vaccines 2019, 7, 197. https://0-doi-org.brum.beds.ac.uk/10.3390/vaccines7040197
Shah AU, Cao Y, Siddique N, Lin J, Yang Q. miR29a and miR378b Influence CpG-Stimulated Dendritic Cells and Regulate cGAS/STING Pathway. Vaccines. 2019; 7(4):197. https://0-doi-org.brum.beds.ac.uk/10.3390/vaccines7040197
Chicago/Turabian StyleShah, Abid Ullah, Yanan Cao, Naila Siddique, Jian Lin, and Qian Yang. 2019. "miR29a and miR378b Influence CpG-Stimulated Dendritic Cells and Regulate cGAS/STING Pathway" Vaccines 7, no. 4: 197. https://0-doi-org.brum.beds.ac.uk/10.3390/vaccines7040197